HATRIC-LRC for small molecules
Now available at Dualsystems
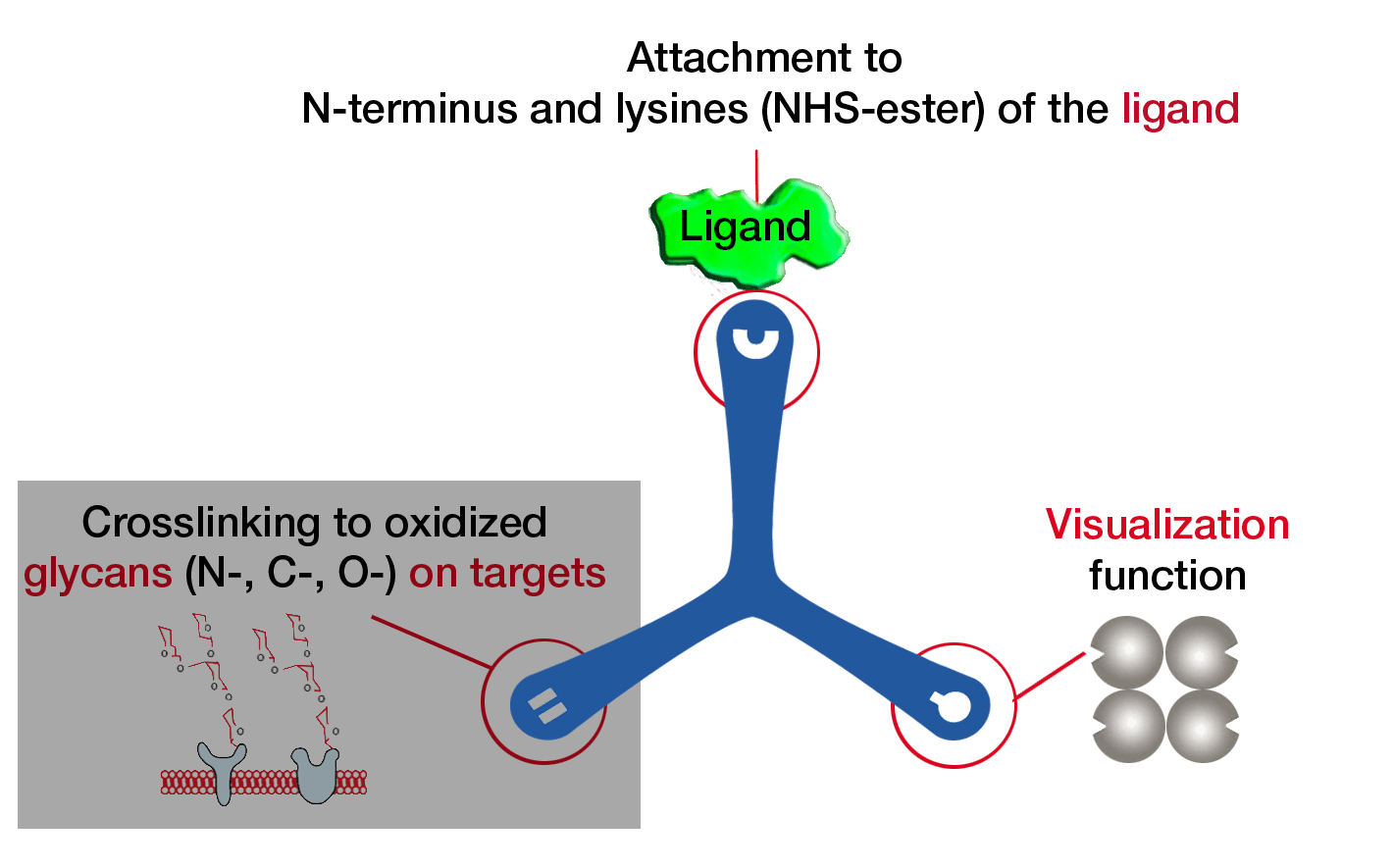
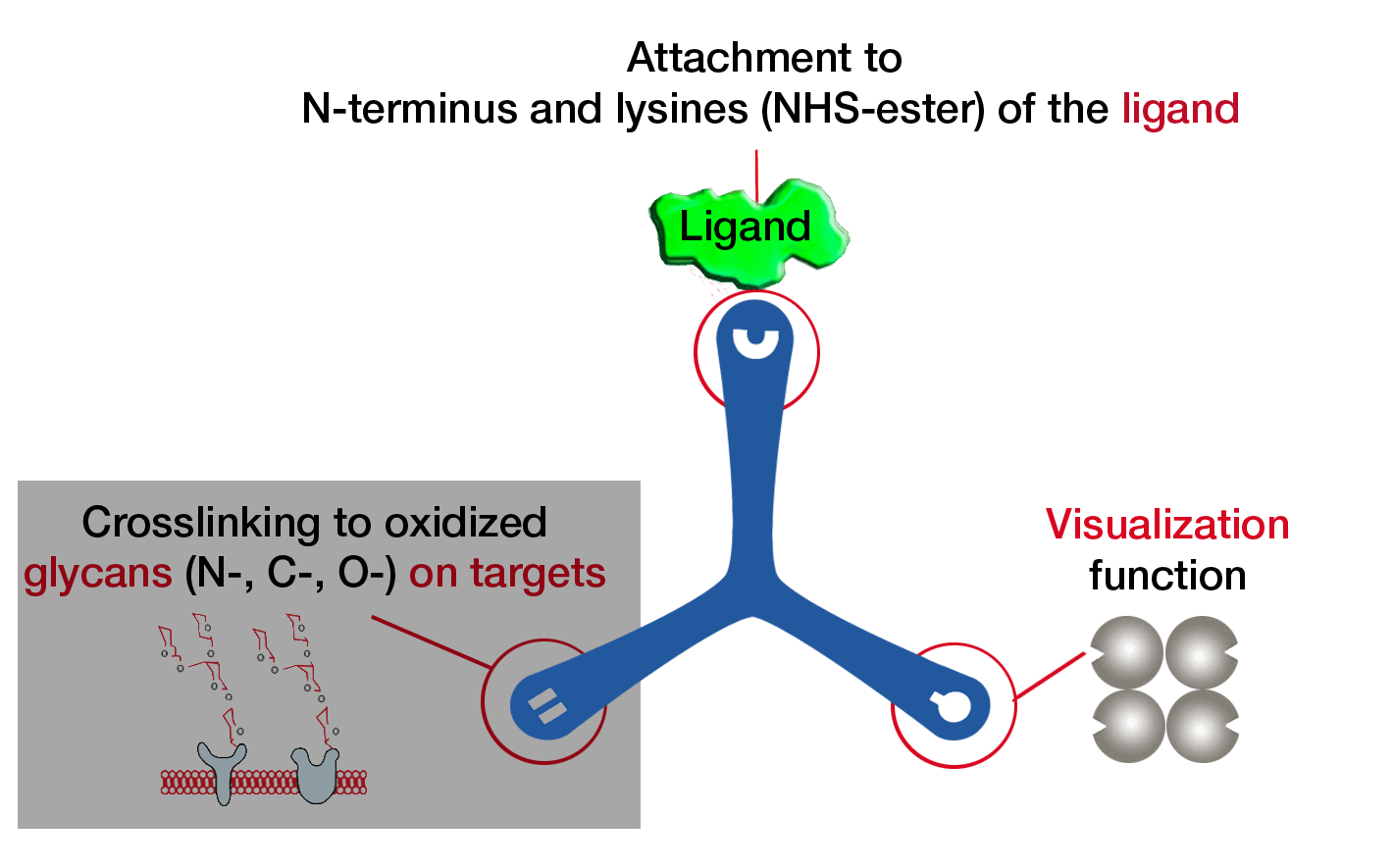
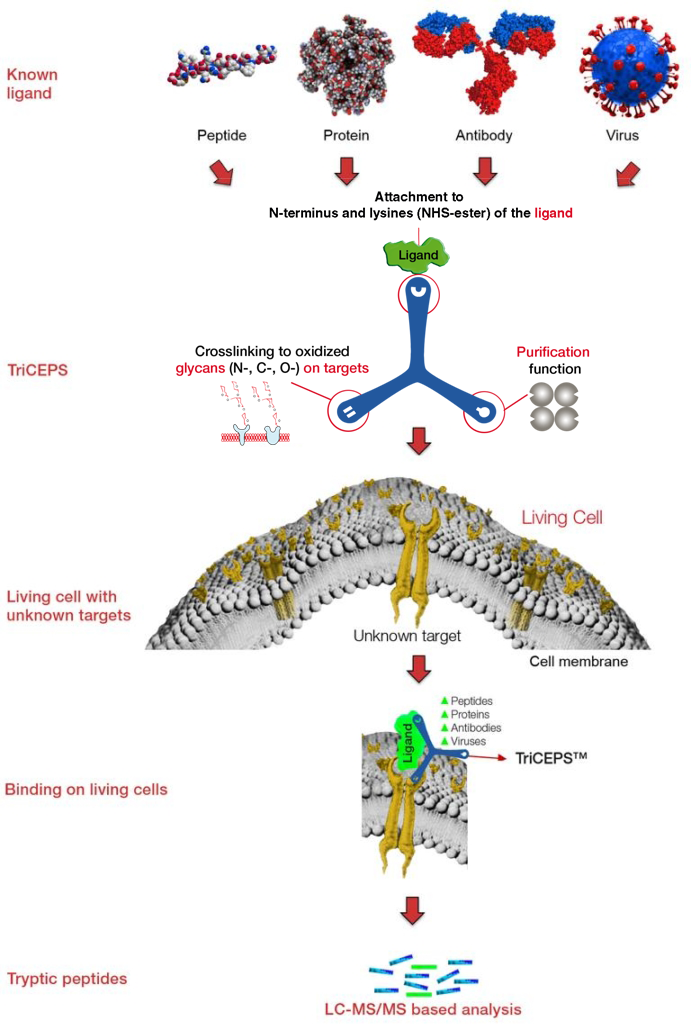
The TriCEPS technology platform has been further developed by the group of Professor Bernd Wollscheid from the ETH Zürich. The newest LRC platform technology is called HATRIC-LRC (HATRIC Ligand Receptor Capture) and enables to identify the targets of small molecule ligands at the cell membrane of living cells. Further, with the HATRIC platform less cells are needed for an experiment than with the original TriCEPS platform and N-, C-, and O-glycosylated targets can be identified.
The HATRIC-platform technology is based on a trifunctional cross linker called HATRIC. One arm of HATRIC contains an N-hydroxysuccinimid (NHS) to be able to bind to small molecules using a primary amine, the second arm contains a hydrazine to covalently bind the glycans of the unknown target proteins at the cell membrane and the third arm possesses an azide to enrich the target proteins.
In a first step the small molecule containing a primary amine is coupled to HATRIC on its first arm. Then the cells expressing the unknown targets and off-targets are mildly oxidized so that aldehydes form on the glycan moieties of the membrane associated proteins. Now, the second arm of HATRIC coupled to the small molecule is able to bind to the glycans at the cell surface. Once the small molecule binds its target the second arm of HATRIC covalently binds to the glycans of the unknown target proteins. In that state the cells expressing the unknown targets are still alive and the target molecules are in their natural microenvironment at the cell membrane. Due to the covalent link the situation is now fixed and the cells are lysed and the third arm of HATRIC is used to enrich the target proteins. The pulled down proteins are then digested with trypsin and identified and relative quantified by LC-MS/MS. Proteins that are enriched in one treatment arm compared to the other treatment arm are the target candidates.


















 De'Broski R. Herbert Ph.D.
De'Broski R. Herbert Ph.D.